2023-02-10
This is Marco's daily open-notebook.
Today is 2023.02.10
Todo today
Theory and ideas of the project
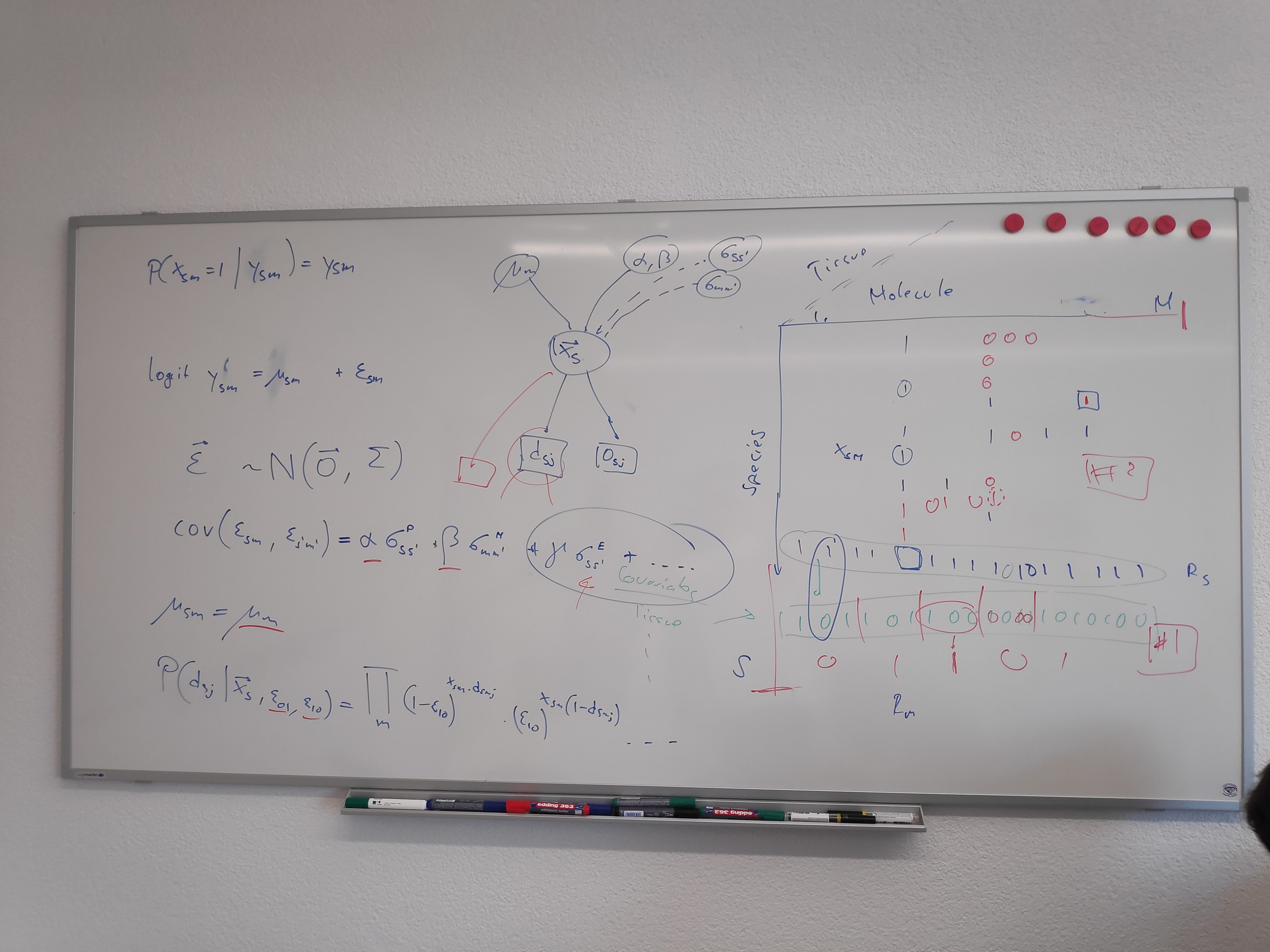
We seek to infer the presence or absence of metabolites in species. We denote by whether metabolite is present () or absent () in species . To infer the full vector , we assume that related species share a similar set of metabolites and that metabolites related in their synthesis share a similar distribution across species. Let be the probability with which metabolite is present in species . We then assume that
where is a metabolite-specific intercept and is normally distributed with mean 0 and co-variance between each combination of species and metabolite. Here, and are known measures of covariance between species and and between metabolites and , respectively, and and are positive scalars.
We consider two sets of data informative about : i) Presence-absence data obtained with mass-spectrometry and ii) presence-only reports of specific metabolites in specific specie. Let be the presence-absence vector of each metabolite obtained with mass-spectrometry run performed on species . Assuming a false-positive and false-negative error rates and , respectively, we have
To model the presence only data, it must be put in relation to the expected research effort. Let denote the known number of presence-only reports for metabolite in species and the unknown number of research projects that aimed at discovering metabolite in species . Assuming a false-positive and false-negative error rates and , respectively, we have
Product of cov ?! and not the sum ?
Images
Here are the first ideas of the discussion we had with Daniel and Pierre-Marie:
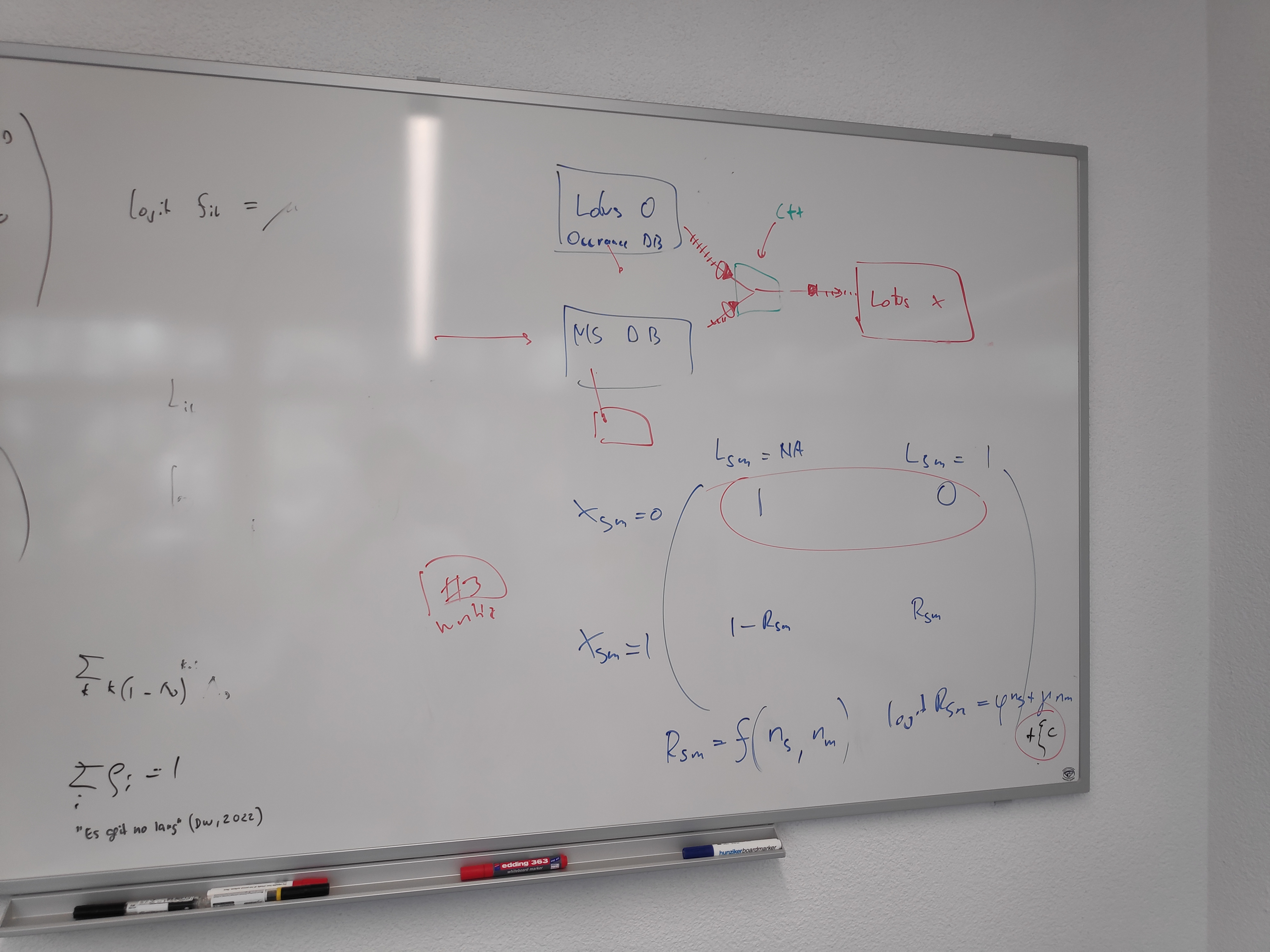
Ideas
See forum discussion here
- The probability of occurence of a given chemical class (i.e of a pseudo absence being indeed pseudo an not a real absence) could be formulated as the sum of probability of this occurence given the : chemical research effort context, phylogenetic context, covariance of other chemical classes.
- This last term was discussed: study of the covariance of chemical classes within a given taxa could be informative.
Doing
Paused
Done
Change prepare_pseudoabsence_tables.R in order to have a csv table and not a space separated data.
Notes
"ref/noref, children/nochildren" mean if you take into account multiple reference (and children) or not.
So, for example: 2 articles report amarogentin in Gentiana lutea, 3 (of which 1 is the same) in Gentiana acaulis.
Then, for the pair amarogentin in Gentiana:
ref_child: 5 → Gives the number of occurences were we find amarogentin related with a Gentiana species.ref_nochild: 4 → Gives how many different papers there are that talk about amarogentin in the species Gentiananoref_child: 2 → Gives how many children of that species were found with at least one reference (in that case : Gentiana
lutea and Gentiana acaulis)noref_nochild: 1 → Will only be either 0 or 1. Gives if it there is at least one reference that talks about that molecule in the species Gentiana
ATTENTION tables are grouped also chemically. So child also refers to the "chemical children", so exactly same principle as for Gentiana and G. lutea but this for Terpenoids > Monoterpenoids > Secoiridoids (> Amarogentin)...